install.packages("jstable")
## From github: latest version
remotes::install_github('jinseob2kim/jstable')
library(jstable)45 亚组分析1行代码实现
前面演示了如何使用purrr进行回归模型的亚组分析及森林图绘制,本来找了好久没找到可以实现这个功能的R包,都打算自己写个包了,没想到这几天找到了!
完美实现COX回归和logistic回归的亚组分析,除此之外,还支持svyglm、svycoxph的结果,并且数据结果可直接用于绘制森林图,连NA和各种空行都给你准备好了!
包的名字叫jstable,直达网址:https://jinseob2kim.github.io/jstable/
这个包除了亚组分析外,还有其他很多函数,大家自己探索即可,我这里就演示下如何进行亚组分析!
45.1 安装
45.2 准备数据
还是使用之前演示的数据。
使用survival包中的colon数据集用于演示,这是一份关于结肠癌患者的生存数据,共有1858行,16列,共分为3个组,1个观察组+2个治疗组,观察他们发生终点事件的差异。
各变量的解释如下:
- id:患者id
- study:没啥用,所有患者都是1
- rx:治疗方法,共3种,Obs(观察组), Lev(左旋咪唑), Lev+5FU(左旋咪唑+5-FU)
- sex:性别,1是男性
- age:年龄
- obstruct:肠梗阻,1是有
- perfor:肠穿孔,1是有
- adhere:和附近器官粘连,1是有
- nodes:转移的淋巴结数量
- status:生存状态,0代表删失,1代表发生终点事件
- differ:肿瘤分化程度,1-well,2-moderate,3-poor
- extent:局部扩散情况,1-submucosa,2-muscle,3-serosa,4-contiguous structures
- surg:手术后多久了,1-long,2-short
- node4:是否有超过4个阳性淋巴结,1代表是
- time:生存时间
- etype:终点事件类型,1-复发,2-死亡
rm(list = ls())
library(survival)
str(colon)
## 'data.frame': 1858 obs. of 16 variables:
## $ id : num 1 1 2 2 3 3 4 4 5 5 ...
## $ study : num 1 1 1 1 1 1 1 1 1 1 ...
## $ rx : Factor w/ 3 levels "Obs","Lev","Lev+5FU": 3 3 3 3 1 1 3 3 1 1 ...
## $ sex : num 1 1 1 1 0 0 0 0 1 1 ...
## $ age : num 43 43 63 63 71 71 66 66 69 69 ...
## $ obstruct: num 0 0 0 0 0 0 1 1 0 0 ...
## $ perfor : num 0 0 0 0 0 0 0 0 0 0 ...
## $ adhere : num 0 0 0 0 1 1 0 0 0 0 ...
## $ nodes : num 5 5 1 1 7 7 6 6 22 22 ...
## $ status : num 1 1 0 0 1 1 1 1 1 1 ...
## $ differ : num 2 2 2 2 2 2 2 2 2 2 ...
## $ extent : num 3 3 3 3 2 2 3 3 3 3 ...
## $ surg : num 0 0 0 0 0 0 1 1 1 1 ...
## $ node4 : num 1 1 0 0 1 1 1 1 1 1 ...
## $ time : num 1521 968 3087 3087 963 ...
## $ etype : num 2 1 2 1 2 1 2 1 2 1 ...可以使用cox回归探索危险因素。分类变量需要变为因子型,这样在进行回归时会自动进行哑变量设置。
为了演示,我们只选择Obs组和Lev+5FU组的患者,所有的分类变量都变为factor,把年龄也变为分类变量并变成factor。
suppressMessages(library(tidyverse))
df <- colon %>%
mutate(rx=as.numeric(rx)) %>%
filter(etype == 1, !rx == 2) %>% #rx %in% c("Obs","Lev+5FU"),
select(time, status,rx, sex, age,obstruct,perfor,adhere,differ,extent,surg,node4) %>%
mutate(sex=factor(sex, levels=c(0,1),labels=c("female","male")),
age=ifelse(age >65,">65","<=65"),
age=factor(age, levels=c(">65","<=65")),
obstruct=factor(obstruct, levels=c(0,1),labels=c("No","Yes")),
perfor=factor(perfor, levels=c(0,1),labels=c("No","Yes")),
adhere=factor(adhere, levels=c(0,1),labels=c("No","Yes")),
differ=factor(differ, levels=c(1,2,3),labels=c("well","moderate","poor")),
extent=factor(extent, levels=c(1,2,3,4),
labels=c("submucosa","muscle","serosa","contiguous")),
surg=factor(surg, levels=c(0,1),labels=c("short","long")),
node4=factor(node4, levels=c(0,1),labels=c("No","Yes")),
rx=ifelse(rx==3,0,1),
rx=factor(rx,levels=c(0,1))
)
str(df)
## 'data.frame': 619 obs. of 12 variables:
## $ time : num 968 3087 542 245 523 ...
## $ status : num 1 0 1 1 1 1 0 0 0 1 ...
## $ rx : Factor w/ 2 levels "0","1": 1 1 2 1 2 1 2 1 1 2 ...
## $ sex : Factor w/ 2 levels "female","male": 2 2 1 1 2 1 2 1 2 2 ...
## $ age : Factor w/ 2 levels ">65","<=65": 2 2 1 1 1 2 2 1 2 2 ...
## $ obstruct: Factor w/ 2 levels "No","Yes": 1 1 1 2 1 1 1 1 1 1 ...
## $ perfor : Factor w/ 2 levels "No","Yes": 1 1 1 1 1 1 1 1 1 1 ...
## $ adhere : Factor w/ 2 levels "No","Yes": 1 1 2 1 1 1 1 1 1 1 ...
## $ differ : Factor w/ 3 levels "well","moderate",..: 2 2 2 2 2 2 2 2 3 2 ...
## $ extent : Factor w/ 4 levels "submucosa","muscle",..: 3 3 2 3 3 3 3 3 3 3 ...
## $ surg : Factor w/ 2 levels "short","long": 1 1 1 2 2 1 1 2 2 1 ...
## $ node4 : Factor w/ 2 levels "No","Yes": 2 1 2 2 2 2 1 1 1 1 ...45.3 亚组分析
使用jstable,只要1行代码即可!!!
library(jstable)
res <- TableSubgroupMultiCox(
# 指定公式
formula = Surv(time, status) ~ rx,
# 指定哪些变量有亚组
var_subgroups = c("sex","age","obstruct","perfor","adhere",
"differ","extent","surg","node4"),
data = df #指定你的数据
)
## Warning in coxph.fit(X, Y, istrat, offset, init, control, weights = weights, :
## Loglik converged before variable 1 ; coefficient may be infinite.
## Warning in coxph.fit(X, Y, istrat, offset, init, control, weights = weights, :
## Loglik converged before variable 1,5,6,7 ; coefficient may be infinite.
res
## Variable Count Percent Point Estimate Lower Upper rx=0 rx=1 P value
## rx Overall 619 100 1.67 1.32 2.11 34.4 48.9 <0.001
## 1 sex <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 2 female 312 50.4 1.32 0.96 1.8 41.1 47.8 0.088
## 3 male 307 49.6 2.29 1.6 3.26 26.6 50.1 <0.001
## 4 age <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 5 >65 224 36.2 1.97 1.33 2.93 29.8 50.6 0.001
## 6 <=65 395 63.8 1.52 1.14 2.03 37 48.1 0.004
## 7 obstruct <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 8 No 502 81.1 1.65 1.27 2.13 34.4 46.8 <0.001
## 9 Yes 117 18.9 1.73 1.01 2.95 34.2 57.6 0.046
## 10 perfor <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 11 No 602 97.3 1.64 1.3 2.08 34.3 48.1 <0.001
## 12 Yes 17 2.7 2.87 0.74 11.21 37.5 77.8 0.129
## 13 adhere <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 14 No 533 86.1 1.69 1.31 2.18 32.9 47.4 <0.001
## 15 Yes 86 13.9 1.5 0.84 2.67 44.4 57.9 0.173
## 16 differ <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 17 well 56 9.2 2.68 1.19 6.02 31 55.6 0.017
## 18 moderate 444 73.3 1.67 1.26 2.21 32 44.8 <0.001
## 19 poor 106 17.5 1.32 0.8 2.19 47.5 64.5 0.277
## 20 extent <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 21 submucosa 18 2.9 0 0 Inf 30 0 0.999
## 22 muscle 70 11.3 2.41 0.93 6.22 9.4 28.9 0.069
## 23 serosa 500 80.8 1.68 1.31 2.16 36.7 52.1 <0.001
## 24 contiguous 31 5 1.44 0.55 3.75 58.4 67.2 0.459
## 25 surg <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 26 short 452 73 1.82 1.37 2.4 31.5 48.9 <0.001
## 27 long 167 27 1.31 0.86 1.99 43.2 49.1 0.208
## 28 node4 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 29 No 453 73.2 1.85 1.37 2.49 27.7 41.5 <0.001
## 30 Yes 166 26.8 1.41 0.97 2.05 53.2 68.7 0.074
## P for interaction
## rx <NA>
## 1 0.029
## 2 <NA>
## 3 <NA>
## 4 0.316
## 5 <NA>
## 6 <NA>
## 7 0.752
## 8 <NA>
## 9 <NA>
## 10 0.442
## 11 <NA>
## 12 <NA>
## 13 0.756
## 14 <NA>
## 15 <NA>
## 16 0.402
## 17 <NA>
## 18 <NA>
## 19 <NA>
## 20 0.1
## 21 <NA>
## 22 <NA>
## 23 <NA>
## 24 <NA>
## 25 0.183
## 26 <NA>
## 27 <NA>
## 28 0.338
## 29 <NA>
## 30 <NA>直接就得出了结果!除了亚组分析的各种结果,还给出了交互作用的P值!
45.4 画森林图
这个结果不需要另存为csv也能直接使用(除非你是细节控,需要修改各种大小写等信息),当然如果你需要HR(95%CI)这种信息,还是需要自己添加一下的。
我们添加个空列用于显示可信区间,并把不想显示的NA去掉即可,还需要把P值,可信区间这些列变为数值型。
plot_df <- res
plot_df[,c(2,3,9)][is.na(plot_df[,c(2,3,9)])] <- " "
plot_df$` ` <- paste(rep(" ", nrow(plot_df)), collapse = " ")
plot_df[,4:6] <- apply(plot_df[,4:6],2,as.numeric)
plot_df
## Variable Count Percent Point Estimate Lower Upper rx=0 rx=1 P value
## rx Overall 619 100 1.67 1.32 2.11 34.4 48.9 <0.001
## 1 sex NA NA NA <NA> <NA>
## 2 female 312 50.4 1.32 0.96 1.80 41.1 47.8 0.088
## 3 male 307 49.6 2.29 1.60 3.26 26.6 50.1 <0.001
## 4 age NA NA NA <NA> <NA>
## 5 >65 224 36.2 1.97 1.33 2.93 29.8 50.6 0.001
## 6 <=65 395 63.8 1.52 1.14 2.03 37 48.1 0.004
## 7 obstruct NA NA NA <NA> <NA>
## 8 No 502 81.1 1.65 1.27 2.13 34.4 46.8 <0.001
## 9 Yes 117 18.9 1.73 1.01 2.95 34.2 57.6 0.046
## 10 perfor NA NA NA <NA> <NA>
## 11 No 602 97.3 1.64 1.30 2.08 34.3 48.1 <0.001
## 12 Yes 17 2.7 2.87 0.74 11.21 37.5 77.8 0.129
## 13 adhere NA NA NA <NA> <NA>
## 14 No 533 86.1 1.69 1.31 2.18 32.9 47.4 <0.001
## 15 Yes 86 13.9 1.50 0.84 2.67 44.4 57.9 0.173
## 16 differ NA NA NA <NA> <NA>
## 17 well 56 9.2 2.68 1.19 6.02 31 55.6 0.017
## 18 moderate 444 73.3 1.67 1.26 2.21 32 44.8 <0.001
## 19 poor 106 17.5 1.32 0.80 2.19 47.5 64.5 0.277
## 20 extent NA NA NA <NA> <NA>
## 21 submucosa 18 2.9 0.00 0.00 Inf 30 0 0.999
## 22 muscle 70 11.3 2.41 0.93 6.22 9.4 28.9 0.069
## 23 serosa 500 80.8 1.68 1.31 2.16 36.7 52.1 <0.001
## 24 contiguous 31 5 1.44 0.55 3.75 58.4 67.2 0.459
## 25 surg NA NA NA <NA> <NA>
## 26 short 452 73 1.82 1.37 2.40 31.5 48.9 <0.001
## 27 long 167 27 1.31 0.86 1.99 43.2 49.1 0.208
## 28 node4 NA NA NA <NA> <NA>
## 29 No 453 73.2 1.85 1.37 2.49 27.7 41.5 <0.001
## 30 Yes 166 26.8 1.41 0.97 2.05 53.2 68.7 0.074
## P for interaction
## rx <NA>
## 1 0.029
## 2 <NA>
## 3 <NA>
## 4 0.316
## 5 <NA>
## 6 <NA>
## 7 0.752
## 8 <NA>
## 9 <NA>
## 10 0.442
## 11 <NA>
## 12 <NA>
## 13 0.756
## 14 <NA>
## 15 <NA>
## 16 0.402
## 17 <NA>
## 18 <NA>
## 19 <NA>
## 20 0.1
## 21 <NA>
## 22 <NA>
## 23 <NA>
## 24 <NA>
## 25 0.183
## 26 <NA>
## 27 <NA>
## 28 0.338
## 29 <NA>
## 30 <NA>
##
## rx
## 1
## 2
## 3
## 4
## 5
## 6
## 7
## 8
## 9
## 10
## 11
## 12
## 13
## 14
## 15
## 16
## 17
## 18
## 19
## 20
## 21
## 22
## 23
## 24
## 25
## 26
## 27
## 28
## 29
## 30画图就非常简单!
library(forestploter)
library(grid)
p <- forest(
data = plot_df[,c(1,2,3,11,9)],
lower = plot_df$Lower,
upper = plot_df$Upper,
est = plot_df$`Point Estimate`,
ci_column = 4,
#sizes = (plot_df$estimate+0.001)*0.3,
ref_line = 1,
xlim = c(0.1,4)
)
print(p)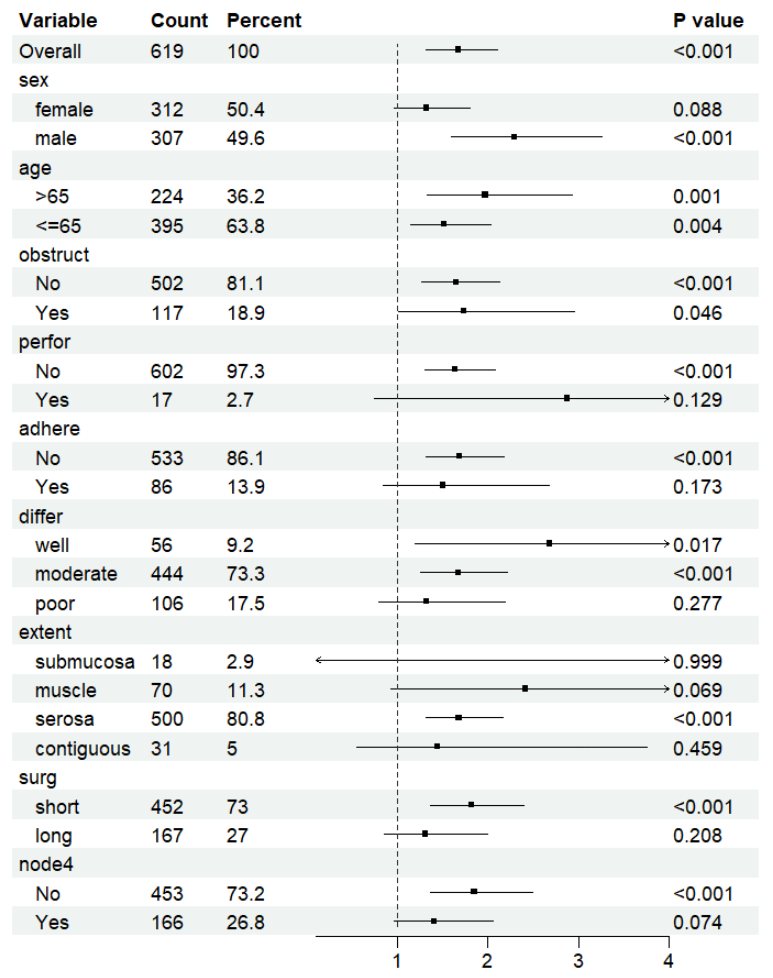
这样就搞定了,真的是非常简单了,省去了大量的步骤。
45.5 其他资源
亚组分析和森林图的内容还有非常多的细节问题,为了不影响该合集的主要内容,我把它们放在下面的链接中,大家感兴趣的话可点击下面的链接查看,或者在公众号后台回复亚组分析获取合集链接: